dsi2 documentation¶
This software is designed to enable the analysis of diffusion-weighted MRI-based tractography datasets. DSI2 stands for D iscrete S patial I ndexing of D iffusion S pectrum I mages. Image reconstruction, tractography and parcellation must be done outside of this software, but indexing, searching and analysis are all implemented here. Specifically, this package provides the tools necessary to perform Local Termination Pattern Analysis (LTPA, [1] )
- The idea is that
- A set of coordinates in the brain can be used to query a database.
- The query returns a set of streamlines that that pass through the set of coordinates.
- The streamlines are divided into groups based on
- Their termination regions
- Clustering based on their shape
- The groups of streamlines are compared across individuals
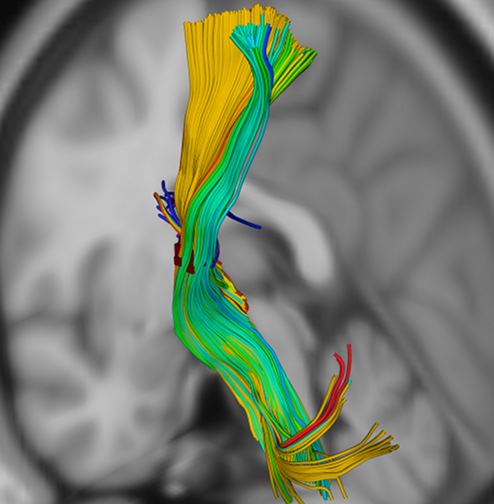
Take, for example, the corticospinal tract. If we select a sphere of coordinates in the internal capsule, we should see tracks following the pathway of the corticospinal tract.
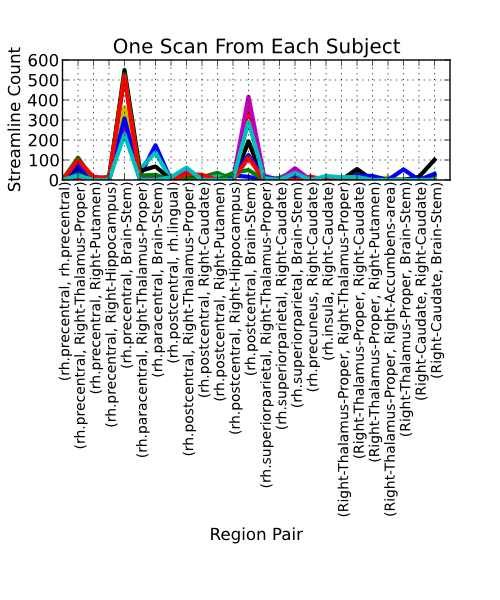
We can then group these streamlines based on the regions in which they terminate. Using the Lausanne2008 scale 33 atlas, we get the following distribution of streamline counts:
Getting Started¶
Preparing data¶
Use DSI Studio and connectome_mapper to prepare data
Organizing your local datasource (Hard way)¶
An overview of possible analyses and pitfalls to beware of.
- Installing DSI2
- DSI2 Download Details
- Preparing data
- Preparing Data with Freesurfer
- DSI2 Reconstruction
- Organizing your local datasource (Hard way)
- Importing your data
- Accessing and analyzing your data
- Creating a streamline aggregator
- Browsing the Streamline Database
- The Sphere Browser: Querying and visualizing streamlines